Michael French
Assistant Professor
Office: CB 151
Education and Training:
- 2000 Bucknell University, B.S. in Cell Biology/Biochemistry
- 2002 Bucknell University, M.S. in Biology; with Kathleen Page
- 2008 Northwestern University, Ph.D. in Biochemistry and Cell/Molecular Biology ; with Linda Hicke
- 2015 Salk Institute for Biological Studies, Postdoc in Biochemistry; with Tony Hunter
- 2018 The Scripps Research Institute, Postdoc in Biochemistry; with Steve Reed
Research Interests:
Research in my lab is focused on the control of protein activity and stability by the ubiquitin system. The ubiquitin system regulates nearly all aspects of eukaryotic cell biology, enabling the modification of thousands of protein targets by a diverse collection of enzymes. Defects in protein ubiquitination have been linked to a number of human diseases, including many cancers and neurodegenerative disorders. We are primarily interested in understanding the biochemical activities of ubiquitin ligases (E3s), which work together with other enzymes to transfer ubiquitin monomers and polymeric ubiquitin chains to protein substrates. The sheer number of E3s (~600 in humans) highlights the vast and extensive nature of regulation by the ubiquitin system.
Specific areas of interest in the lab include understanding the structures and functions of unconventional ubiquitin chain modifications (French et al., 2021; Gregor et al., 2023) as well as the molecular mechanisms underlying their synthesis (French et al., 2017). We aim to elucidate the architectures and biological roles of these "heterotypic" (mixed and branched; Fig. 1A) ubiquitin chains using a combination of biochemistry, cell/molecular biology, and proteomics. We are also interested in understanding how the activities of HECT E3s (e.g., ITCH and UBR5; Fig. 1B), the second largest class of ubiquitin ligases, are controlled and fine-tuned to ensure the fidelity of protein ubiquitination.
Please contact me at mfrench@ut.edu for further information about specific research projects and opportunities in my lab.
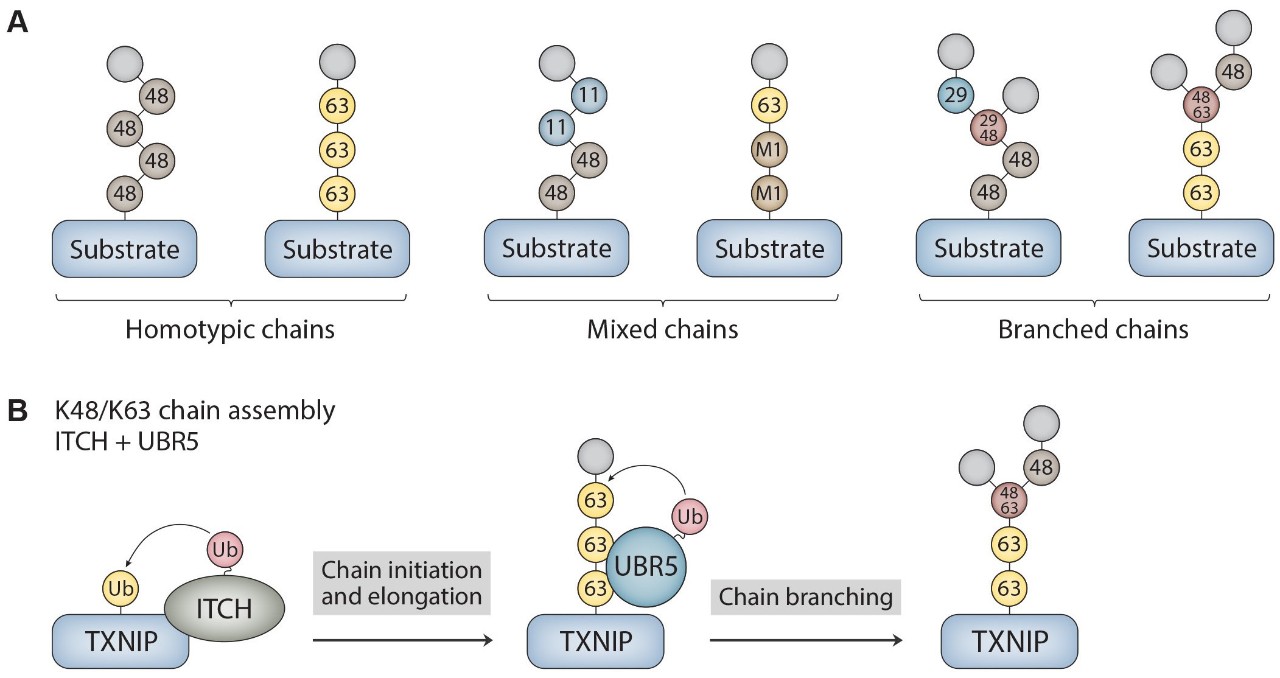
Figure 1. Ubiquitin chain architectures and mechanism of branched K48/K63 chain assembly. (A) Ubiquitin chains can be classified as either homotypic or heterotypic (mixed and branched) based on the types of ubiquitin linkages and how the subunits are connected to each other within the chain. Examples of homotypic K48- and K63-linked chains are shown. Mixed K11/K48 and M1/K63 chains as well as branched K29/K48 and K48/K63 chains are shown as examples of heterotypic chains. (B) Mechanism of branched chain formation by the HECT ubiquitin ligases ITCH and UBR5. ITCH first attaches homotypic K63-linked chains to TXNIP. UBR5 then binds to the K63 linkages through its UBA domain to nucleate the formation of K48 linkages, resulting in the assembly of branched K48/K63 chains
Biochemistry is an inherently interdisciplinary field of study. Students in my lab use a variety of techniques and experimental approaches, including protein purification, in vitro enzyme assays, analytical protein/peptide techniques, mass spectrometry (collaborative), and yeast cell biology. All students work on original research projects and are involved in each stage of the scientific process: planning, executing, analyzing, presenting and writing. I expect my students to build on the concepts and techniques that they were introduced to in their courses and to develop their scientific skills through participation in hands-on research, group meetings, journal club discussions, oral presentations and grant/paper writing.
Gregor, J.B.; Xu, D.; French, M.E.* "Assembly and disassembly of branched ubiquitin chains." Front. Mol. Biosci., 2023, 10, 1-7.
French, M.E.*; Koehler, C.F.; Hunter, T.* "Emerging functions of branched ubiquitin chains." Cell Discov., 2021, 7, 1-10.
French, M.E.; Klosowiak, J.L.; Aslanian, A.; Reed, S.I.; Yates, J.R. III; Hunter, T.* "Mechanism of ubiquitin chain synthesis employed by a HECT E3." J. Biol. Chem., 2017, 292, 10398-10413.
Klosowiak, J.L.; Park, S.; Smith, K.P.; French, M.E.; Focia, P.J.; Freymann, D. M.; Rice, S.E.* "Structural insights into Parkin substrate lysine targeting from minimal Miro substrates." Sci. Rep., 2016, 6, 1-13.
French, M.E.; Kretzmann, B.R.; Hicke, L.* "Regulation of the Rsp5 ubiquitin ligase by an intrinsic ubiquitin-binding site." J. Biol. Chem., 2009, 284, 12071-12079.
Stamenova, S.D.; French, M.E.; He., Y.; Francis, S.A.; Kramer, Z.B.; Hicke, L.* "Ubiquitin binds to and regulates a subset of SH3 domains." Mol. Cell, 2007, 25, 273-284.
French, M.E.; Swanson, K.; Shih, S.C.; Radhakrishnan, I.; Hicke, L.* "Identification and characterization of modular domains that bind ubiquitin." Methods Enymol., 2005, 399, 135-157.

